Life Science Institute
(684 Sec. 1, Chung Hua Rd. Taitung, Taiwan. 950)
Text: Lewin, B. 2005 Genes VIII. Oxford, New York
Lecturer:Dr. Lee, Yen
My phone number is(886—89—318855 ext:2531, and my e-mail address is : yenlee@nttu.edu.tw )
I will use lot of online open to public visual teaching resources to explain theories. I have to say thanks for those kind people providing those free tools. This course is provided for free.
Course Outline:
Chapter 1. Introduction
Let us watch this movie :
http://science.nhmccd.edu/biol/bio1int.htm#genetic
Exploring Our Molecular Selves" Video
The basics:
The hereditary of a living organism is defined by its genome, which constructed of nucleic acids.
Let's check the following web resources:
http://www.pbs.org/wgbh/nova/genome/dna_sans.html
http://www.sciencenetlinks.com/interactives/dna.swf
The structure of double strand DNA:
http://www.biostudio.com/demo_freeman_dna_coiling.htm
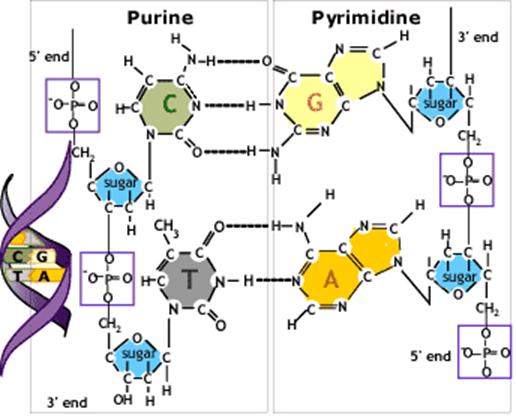
DNA stands for deoxyribonucleic acid which is a structure of sugar, phosphate and a base combined into a complex double helix. The two strands in the helix are complementary to each other, which means that each DNA strand contains the template information for synthesis of a new copy of the other strand.
The building blocks of DNA are the 5-carbon sugar deoxyribose linked together by phosphodiester bonds forming two strands of sugar-phosphate backbones on the outside of the double helix. Each ribose also binds one of four alternative bases: adenine (A), guanine (G), cytosine (C) or thymine (T).The opposing strands are held together by basepairing between the two strands: G is always paired with C by three hydrogen bonds and A is always paired with T by two hydrogen bonds.
Let’s check the DNA structure again:

(http://nobelprize.org/educational_games/medicine/dna/b/replication/dna_structure.html)
A gene is a sequence of DNA that codes for an RNA; in protein-coding genes, the RNA in turn codes for a protein.
How do we know DNA is the genetic material?
The first hint:
The discovery of transformation :
http://life.nthu.edu.tw/~rrandd/90s1/b881622/bioinfo.swf
Purification of the transforming principle in 1944 showed that it is deoxyribonucleic acid (DNA).
Second Hint:
Hershey and Chase Experiment
http://highered.mcgraw-hill.com/olc/dl/120076/bio21.swf
According to the above experiments, we know DNA is the genetic material.
The basic building block of nucleic acids is the nucleotide. It has three components:a nitrogenous base; a sugar; and a phosphate
To differentiate between the numbering systems of the heterocyclic rings and the sugar, positions on the pentose are given a prime (′).
Nucleic acids are named for the type of sugar; DNA has 2′–deoxyribose, whereas RNA has ribose. The difference is that the sugar in RNA has an OH group at the 2′ position of the pentose ring.
In RNA uracil is found instead of thymine.
The terminal nucleotide at one end of the chain has a free 5′ group; the terminal nucleotide at the other end has a free 3′ group. We usually write nucleic acid sequences in the 5′→3′ direction.
http://www.pwc.k12.nf.ca/wadey/biotech/dna1.swf
http://www.sciencetechnologies.com/examples/example7/example7/index.html
DNA Chemical Properties
The process of strand separation is called denaturation or melting.
The midpoint of the temperature range over which the strands of DNA separate is called the melting temperature (Tm).
How to calculate the melting temperature:
http://www.psb.ugent.be/rRNA/primers/tm.html
The ability of the two separated complementary strands to reform into a double helix is called renaturation.
The technique to allow any two complementary nucleic acid sequences to react with each other to form a duplex structure is sometimes called annealing, or hybridization
http://highered.mcgraw-hill.com/olc/dl/120078/bio_g.swf
DNA Replication:
Semiconservative replication:
Matthew Meselson and Frank W. Stahl experiment:
http://highered.mcgraw-hill.com/olc/dl/120076/bio22.swf
http://highered.mcgraw-hill.com/olc/dl/120076/micro04.swf
http://highered.mcgraw-hill.com/olc/dl/120076/bio23.swf
http://www.wiley.com/college/pratt/0471393878/student/animations/dna_replication/index.html
http://bcs.whfreeman.com/thelifewire/content/chp11/1102003.html
Mutation:
http://highered.mcgraw-hill.com/olc/dl/120082/bio36.swf
http://highered.mcgraw-hill.com/olc/dl/120082/bio33.swf
http://highered.mcgraw-hill.com/olc/dl/120082/bio32.swf
http://highered.mcgraw-hill.com/olc/dl/120082/micro18.swf
http://www.accessexcellence.org/RC/VL/GG/mutation2.html
http://www.accessexcellence.org/RC/VL/GG/mutation.html
http://www.ucl.ac.uk/~sjjgsca/DNAmutation.html
http://www.emunix.emich.edu/~rwinning/genetics/mutat2.htm
http://www.tokyo-med.ac.jp/genet/dmu-e.htm
Diseases caused by genetic mutations:
The end of chapter one.
Chapter 2. Genetic Information Flow
The central dogma:
DNA==(transcription)=>RNA(mRNA tRNA rRNA)==(translation)=>Protein
Let’s check these visual teaching aids:
http://highered.mcgraw-hill.com/olc/dl/120077/bio25.swf
http://nobelprize.org/educational_games/medicine/dna/intro.html
http://nobelprize.org/educational_games/medicine/dna/b/transcription/transcription_ani.html
http://www.fed.cuhk.edu.hk/~johnson/teaching/genetics/animations/transcription.htm
http://www.sumanasinc.com/webcontent/anisamples/molecularbiology/polyribosomes.html
We call these procedures=>The Central Dogma of molecular biology.
Let’s check more visual teaching aids:
http://www.accessexcellence.org/RC/VL/GG/central.html
http://207.207.4.198/pub/flash/26/transmenu_s.swf
http://web.mit.edu/esgbio/www/dogma/dogma.html
http://www.cbs.dtu.dk/staff/dave/DNA_CenDog.html
RNA Structure
http://www-scf.usc.edu/~chem203/resources/DNA/rna_structure.html
http://www.web-books.com/MoBio/Free/Ch3C.htm
Transcription and Translation
Transcription:
http://www.csuchico.edu/~jbell/Biol207/animations/transcription.html
http://www-class.unl.edu/biochem/gp2/m_biology/animation/gene/gene_a2.html
http://vcell.ndsu.nodak.edu/animations/transcription/movie.htm
http://library.thinkquest.org/20465/g_DNATranscription.html
Translation:
http://www.ncc.gmu.edu/dna/ANIMPROT.htm
http://nobelprize.org/medicine/educational/dna/b/translation/translation_ani.html
http://highered.mcgraw-hill.com/olc/dl/120077/micro06.swf
http://www.biostudio.com/demo_freeman_protein_synthesis.htm
http://telstar.ote.cmu.edu/Hughes/HughesArchive/tutorial/polypeptide/tutorial.swf
http://www.wisc-online.com/objects/index_tj.asp?objid=AP1302
Genetic Codes
http://psyche.uthct.edu/shaun/SBlack/geneticd.html
http://www.accessexcellence.org/RC/VL/GG/genetic.html
http://web.indstate.edu/thcme/mwking/protein-synthesis.html
http://cellbio.utmb.edu/cellbio/ribosome.htm
http://bcs.whfreeman.com/thelifewire/content/chp12/1202002.html
Every sequence has three possible reading frames.
The Direction of Protein Synthesis:
http://www.johnkyrk.com/aminoacid.html
 (http://web.mit.edu/esgbio/www/lm/proteins/peptidebond.html
)
(http://web.mit.edu/esgbio/www/lm/proteins/peptidebond.html
)
NH2èCOOH
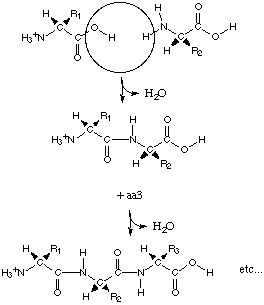
Protein structure:
http://www.rothamsted.ac.uk/notebook/courses/guide/prot.htm
http://www.ausetute.com.au/proteins.html
Enzymes are proteins:
Enzyme function:
http://www.stolaf.edu/people/giannini/flashanimat/enzymes/prox-orien.swf
http://www.tvdsb.on.ca/westmin/science/sbi3a1/digest/enzymes.htm
http://programs.northlandcollege.edu/biology/Biology1111/animations/glycolysis.html
Message RNA Processing --Introns and Extrons
http://www.sumanasinc.com/webcontent/anisamples/molecularbiology/mRNAsplicing.html http://highered.mcgraw-hill.com/olc/dl/120077/bio30.swf
http://vcell.ndsu.nodak.edu/animations/mrnaprocessing/movie.htm
http://bcs.whfreeman.com/thelifewire/content/chp14/1402001.html
DNA Research Tools
Restriction endonucleases are a key tool in manipulating DNA
http://highered.mcgraw-hill.com/olc/dl/120078/bio37.swf
http://highered.mcgraw-hill.com/olc/dl/120078/bio38.swf
http://www.fhcrc.org/science/education/hs/hutchlab/lessons/animate/ecorv.html
http://www.dnai.org/text/mediashowcase/index2.html?id=549
http://learn.genetics.utah.edu/units/biotech/index.cfm
http://www.sumanasinc.com/webcontent/anisamples/molecularbiology/plasmidcloning_fla.html
http://highered.mcgraw-hill.com/olc/dl/120078/micro10.swf
Life cycle of mRNAs:
http://www.sumanasinc.com/webcontent/anisamples/molecularbiology/lifecyclemrna_fla.html
Exon sequences are conserved but introns vary.
Some DNA sequences code for more than one protein.This situation arises when the same sequence of DNA is translated in more than one reading frame. In cellular genes, a DNA sequence usually is read in only one of the three potential reading frames, but in some viral and mitochondrial genes, there is an overlap between two adjacent genes that are read in different reading frames.
In some genes, alternative patterns of gene expression create switches in the pathway for connecting the exons. A single gene may generate a variety of mRNA products that differ in their content of exons. The difference may be that certain exons are optional—they may be included or spliced out. Sometimes the pathways are alternatives that are expressed under different conditions, one in one cell type and one in another cell type.
http://www.learner.org/channel/courses/biology/units/proteo/images.html
Other research tools:
Electrophoresis:
http://bcs.whfreeman.com/thelifewire/content/chp16/1602001.html
Micro array:
http://bcs.whfreeman.com/thelifewire/content/chp16/1602002.html
DNA testing by allele-specific cleavage:
http://bcs.whfreeman.com/thelifewire/content/chp17/1702001.html
The end of chapter two.
Chapter 3. Genome
The genome is the complete set of DNA sequences in an organism.
How we sequence the genome:
DNA sequencing method:
http://www.biostudio.com/case_freeman_dna_sequencing.html
http://www.dnai.org/text/mediashowcase/index2.html?id=608
http://smcg.cifn.unam.mx/enp-unam/03-EstructuraDelGenoma/animaciones/secuencia.swf
The transcriptome is the complete set of RNAs present in a cell, tissue, or organism.
The proteome is the complete set of proteins that is expressed by the entire genome.
The coexistence of multiple alleles at a locus is called genetic polymorphism.
A change in a single nucleotide when alleles are compared is called a single nucleotide polymorphism (SNP).
A difference in restriction maps between two individuals is called a restriction fragment length polymorphism (RFLP).
http://highered.mcgraw-hill.com/olc/dl/120078/bio20.swf
RFLPs and SNPs can be used for genetic mapping.
If we compare the restriction map of DNA from patients suffering from a disease with the DNA of normal people, we may find that a particular restriction site is always present (or always absent) from the patients. It would imply that the restriction marker lies so close to the mutant gene that it is never separated from it by recombination.
The use of DNA restriction analysis to identify individuals has been called DNA fingerprinting.
http://www.pbs.org/wgbh/nova/sheppard/analyze.html
http://protist.biology.washington.edu/fingerprint/dnaintro.html
http://www.thenakedscientists.com/HTML/Columnists/dalyacolumn8.htm
http://www.pbs.org/wgbh/nova/sheppard/labwave.html
Eukaryotic genomes contain both nonrepetitive and repetitive DNA sequences
Repetitive DNA is often divided into two types:
Moderately repetitive DNA consists of relatively short sequences that are repeated typically 10-1000x in the genome.
Highly repetitive DNA consists of very short sequences (typically <100 bp) that are present many thousands of times in the genome, often organized as long tandem repeats
Neither class represents protein.
A significant part of the moderately repetitive DNA consists of transposons, short sequences of DNA (~1 kb) that have the ability to move to new locations in the genome and/or to make additional copies of themselves
All of the bacteria with genome sizes below 1.5 Mb are obligate intracellular parasites
The free-living bacterium with the smallest known genome is the thermophile Aquifex aeolicus, with 1.5 Mb and 1512 genes (2373). A "typical" gram-negative bacterium, H. influenzae, has 1,743 genes each of ~900 bp. So we can conclude that ~1500 genes are required to make a free-living organism.
Eukaryotic genes are transcribed individually, each gene producing a monocistronic messenger with few exceptions.
An active gene should consist of a series of exons where the first exon immediately follows a promoter, the internal exons are flanked by appropriate splicing junctions, the last exon is followed by 3′ processing signals, and a single open reading frame starting with an initiation codon and ending with a termination codon can be deduced by joining the exons together.
Repetitive sequences account for >50% of the human genome. The repetitive sequences fall into five classes:
Transposons (either active or inactive) account for the vast majority (45% of the genome). All transposons are found in multiple copies.
Processed pseudogenes (~3000 in all, account for ~0.1% of total DNA).
Simple sequence repeats (highly repetitive DNA such as (CA)n account for ~3%).
Segmental duplications (blocks of 10-300 kb that have been duplicated into a new region) account for ~5%.
Tandem repeats form blocks of one type of sequence (especially found at centromeres and telomeres).
Genes are expressed at widely differing levels
The proportion of DNA represented in an mRNA population can be determined by the amount of the DNA that can hybridize with the RNA. Such a saturation analysis typically identifies ~1% of the DNA as providing a template for mRNA.
Many somatic tissues of higher eukaryotes have an expressed gene number in the range of 10,000-20,000.
The expressed gene number of chick liver is ~11,000-17,000, compared with the value for oviduct of ~13,000-15,000. How many of these two sets of genes are identical?
How many are specific for each tissue? These questions are usually addressed by analyzing the transcriptome—the set of sequences represented in RNA.
For example, ~75% of the sequences expressed in liver and oviduct are the same. In other words, ~12,000 genes are expressed in both liver and oviduct, ~5000 additional genes are expressed only in liver, and ~3000 additional genes are expressed only in oviduct. Between mouse liver and kidney, ~90% of the mRNAs are identical, leaving a difference between the tissues of only 1000-2000 in terms of the number of expressed genes.
Only ~10% of the mRNA sequences of a cell are unique to it. The majority of sequences are common to many, perhaps even all, cell types.
This suggests that the common set of expressed gene functions, numbering perhaps ~10,000 in a mammal, comprise functions that are needed in all cell types. Sometimes this type of function is referred to as a housekeeping gene or constitutive gene.
The name microsatellite is used when the length of the repeating unit is <10 bp, and the name minisatellite is used when the length of the repeating unit is ~10-100 bp. These types of sequences are also called VNTR (variable number tandem repeat) regions.
The
high variability of minisatellites makes them especially useful for genomic
mapping, because there is a high probability that individuals will vary in their
alleles at such a locus.
The end of chapter 3.
Chapter 4. Messenger RNA, tRNA, rRNA, Protein Synthesis
Three major classes of RNA are directly involved in the production of proteins.
The following animation takes a little bit time to load:
http://www3.interscience.wiley.com:8100/legacy/college/boyer/0471661791/structure/tRNA/trna.htm
More animations to explain transcription:
http://www.johnkyrk.com/DNAtranscription.html
http://purana.csa.iisc.ernet.in/~mbk/visual.html
http://www.phschool.com/science/biology_place/biocoach/transcription/mrnaeuk.html
http://140.116.60.1/mdlai/Handout/transcription-med-99/sld001.htm
http://www.tokyo-med.ac.jp/genet/dmp-e.htm
http://student.ccbcmd.edu/courses/bio141/lecguide/unit1/prostruct/transcription/transc.html
More animations to explain translation:
http://student.ccbcmd.edu/~gkaiser/biotutorials/protsyn/init.html
http://student.ccbcmd.edu/~gkaiser/biotutorials/protsyn/50s.html
http://student.ccbcmd.edu/~gkaiser/biotutorials/protsyn/peptidea.html
http://student.ccbcmd.edu/~gkaiser/biotutorials/protsyn/stop.html
http://student.ccbcmd.edu/~gkaiser/biotutorials/protsyn/translat.html
http://www.johnkyrk.com/DNAtranslation.html
http://www.biostudio.com/demo_freeman_protein_synthesis.htm
http://carbon.cudenver.edu/~bstith/transla.MOV
http://telstar.ote.cmu.edu/Hughes/HughesArchive/tutorial/polypeptide/tutorial.swf
http://www.execulink.com/~ekimmel/translate.htm
http://www.mun.ca/biochem/courses/3107/Lectures/Topics/Ingredients.html [*]
http://biology.kenyon.edu/courses/biol114/Chap05/Chapter05.html [*]
http://biology.kenyon.edu/slonc/bio3/ribo/ribo1.html
http://plato.acadiau.ca/courses/biol/Microbiology/Translation.htm
mRNA modification
http://www.zerobio.com/mrna_flash.htm
http://employees.csbsju.edu/hjakubowski/classes/ch331/dna/oldnacentdogma.html
A protein-synthesizing system from one cell type can translate the mRNA from another, demonstrating that both the genetic code and the translation apparatus are universal.
All tRNAs have common secondary and tertiary structures. The tRNA secondary structure can be written in the form of a cloverleaf.
The overall range of tRNA lengths is 74-95 bases. The variation in length is caused by differences in the D arm and extra arm.
When a tRNA is charged with the amino acid corresponding to its anticodon, it is called aminoacyl-tRNA. The amino acid is linked by an ester bond from its carboxyl group to the 2′ or 3′ hydroxyl group of the ribose of the 3′ terminal base of the tRNA (which is always adenine). The process of charging a tRNA is catalyzed by a specific enzyme, aminoacyl-tRNA synthetase.
There are (at least) 20 aminoacyl-tRNA synthetases. Each recognizes a single amino acid and all the tRNAs on to which it can legitimately be placed.
http://www.uvm.edu/~dstratto/bcor011_handouts/28_translation2.ppt#16
There is at least one tRNA (but usually more) for each amino acid. A tRNA is named by using the three letter abbreviation for the amino acid as a superscript. If there is more than one tRNA for the same amino acid, subscript numerals are used to distinguish them. So two tRNAs for tyrosine would be described as tRNA1Tyr and tRNA2Tyr. A tRNA carrying an amino acid—that is, an aminoacyl-tRNA—is indicated by a prefix that identifies the amino acid. Ala-tRNA describes tRNAAla carrying its amino acid.
http://www.dnai.org/text/mediashowcase/index2.html?id=586
Translation of an mRNA into a polypeptide chain is catalyzed by the ribosome. Ribosomes are traditionally described in terms of their (approximate) rate of sedimentation (measured in Svedbergs, in which a higher S value indicates a greater rate of sedimentation and a larger mass). Bacterial ribosomes generally sediment at ~70S. The ribosomes of the cytoplasm of higher eukaryotic cells are larger, usually sedimenting at ~80S.
Bacterial (70S) ribosomes have subunits that sediment at 50S and 30S. The subunits of eukaryotic cytoplasmic (80S) ribosomes sediment at 60S and 40S.
The 30S subunit of each ribosome is associated with the mRNA, and the 50S subunit carries the newly synthesized protein. The tRNA spans both subunits.
http://alf1.mrc-lmb.cam.ac.uk/~ribo/movies/
Bacterial mRNA usually is unstable, and is therefore translated into proteins for only a few minutes.
In a eukaryotic cell, synthesis and maturation of mRNA occur in the nucleus. After these events are completed,the mRNA exported to the cytoplasm, where it is translated by ribosomes. Eukaryotic mRNA is relatively stable and continues to be translated for several hours.
Bacterial transcription and translation take place at similar rates. At 37°C, transcription of mRNA occurs at ~40 nucleotides/second. This is very close to the rate of protein synthesis, roughly 15 amino acids/second. It therefore takes ~2 minutes to transcribe and translate an mRNA of 5000 bp, corresponding to 180 kD of protein. When expression of a new gene is initiated, its mRNA typically will appear in the cell within ~2.5 minutes. The corresponding protein will appear within perhaps another 0.5 minute.
The stability of mRNA has a major influence on the amount of protein that is produced. It is usually expressed in terms of the half-life. The mRNA representing any particular gene has a characteristic half-life, but the average is ~2 minutes in bacteria.
Bacterial mRNAs vary greatly in the number of proteins for which they code. Some mRNAs represent only a single gene: they are monocistronic. Others (the majority) carry sequences coding for several proteins: they are polycistronic. In these cases, a single mRNA is transcribed from a group of adjacent genes. (Such a cluster of genes constitutes an operon that is controlled as a single genetic unit;
http://www.exonhit.com/UserFiles/Image/epissage.swf?PHPSESSID=6c8q4ebftvbfh6kfcv96e202e4
All mRNAs contain two types of region. The coding region consists of a series of codons representing the amino acid sequence of the protein, starting (usually) with AUG and ending with a termination codon. But the mRNA is always longer than the coding region, extra regions are present at both ends. An additional sequence at the 5′ end, preceding the start of the coding region, is described as the leader or 5′ UTR (untranslated region). An additional sequence following the termination signal, forming the 3′ end, is called the trailer or 3′ UTR. Although part of the transcription unit, these sequences are not used to code for protein.
http://brodylab.eng.uci.edu/cgi-bin/jpbrody/animation/files/19-981600662.html
http://brodylab.eng.uci.edu/cgi-bin/jpbrody/animation/files/3-973824759.html
The first methylation occurs in all eukaryotes, and consists of the addition of a methyl group to the 7 position of the terminal guanine. A cap that possesses this single methyl group is known as a cap 0. This is as far as the reaction proceeds in unicellular eukaryotes. The enzyme responsible for this modification is called guanine-7-methyltransferase.
The next step is to add another methyl group, to the 2′–O position of the penultimate base(which was actually the original first base of the transcript before any modifications were made). This reaction is catalyzed by another enzyme (2′–O-methyl-transferase). A cap with the two methyl groups is called cap 1. This is the predominant type of cap in all eukaryotes except unicellular organisms.
http://student.ccbcmd.edu/~gkaiser/biotutorials/protsyn/exon.html
http://vcell.ndsu.nodak.edu/animations/mrnaprocessing/movie.htm
http://www.biologie.uni-hamburg.de/b-online/library/bio201/mrnacap.html
http://www.biologie.uni-hamburg.de/b-online/library/bio201/rnasplice.html
http://www.tracy.k12.ca.us/thsadvbio/animations/ProteinSynthesis.swf
In a small minority of cases in higher eukaryotes, another methyl group is added to the second base. This happens only when the position is occupied by adenine; the reaction involves addition of a methyl group at the N6 position. The enzyme responsible acts only on an adenosine substrate that already has the methyl group in the 2′–O position.
In some species, a methyl group is added to the third base of the capped mRNA. The substrate for this reaction is the cap 1 mRNA that already possesses two methyl groups. The third-base modification is always a 2′–O ribose methylation. This creates the cap 2 type. This cap usually represents less than 10-15% of the total capped population.
The poly(A) sequence is not coded in the DNA, but is added to the RNA in the nucleus after transcription. The addition of poly(A) is catalyzed by the enzyme poly(A) polymerase, which adds ~200 A residues to the free 3′-OH end of the mRNA. The poly(A) tract of both nuclear RNA and mRNA is associated with a protein, the poly(A)-binding protein (PABP).
The poly(A) region of mRNA can base pair with oligo(U) or oligo(dT); and this reaction can be used to isolate poly(A)+ mRNA.
RNAase E is the key enzyme in initiating cleavage of mRNA. It may be the enzyme that makes the first cleavage for many mRNAs.
RNAase E was originally discovered as the enzyme that is responsible for processing 5′ rRNA from the primary transcript by a specific endonucleolytic processing event.
The process of degradation may be catalyzed by a multienzyme complex (sometimes called the degradosome) that includes ribonuclease E, PNPase, and a helicase (970). RNAase E plays dual roles. Its N-terminal domain provides an endonuclease activity. The C-terminal domain provides a scaffold that holds together the other components. The helicase unwinds the substrate RNA to make it available to PNPase.
The AUG codon represents methionine, and two types of tRNA can carry this amino acid. One is used for initiation, the other for recognizing AUG codons during elongation.
In bacteria and in eukaryotic organelles, the initiator tRNA carries a methionine residue that has been formylated on its amino group, forming a molecule of N-formyl-methionyl-tRNA. The tRNA is known as tRNAfMet.
(What is formylation?
=>http://en.wikipedia.org/wiki/Formylation_reaction )
This tRNA is used only for initiation. It recognizes the codons AUG or GUG (occasionally UUG). The codons are not recognized equally well: the extent of initiation declines about half when AUG is replaced by GUG, and declines by about half again when UUG is employed.
tRNAmMet. This tRNA responds only to internal AUG codons. Its methionine cannot be formylated.
The initiation sequences protected by bacterial ribosomes are ~30 bases long. The ribosome-binding sites of different bacterial mRNAs display two common features:
The AUG (or less often, GUG or UUG) initiation codon is always included within the protected sequence.
Within 10 bases upstream of the AUG is a sequence that corresponds to part or all of the hexamer.
5′ ... A G G A G G ... 3′
This polypurine stretch is known as the Shine-Dalgarno sequence. It is complementary to a highly conserved sequence close to the 3′ end of 16S rRNA.
Written in reverse direction, the rRNA sequence is the hexamer:
3′ ... U C C U C C ... 5′
The sequence at the 3′ end of rRNA is conserved between prokaryotes and eukaryotes except that in all eukaryotes there is a deletion of the five-base sequence CCUCC that is the principal complement to the Shine-Dalgarno sequence. There does not appear to be base pairing between eukaryotic mRNA and 18S rRNA. This is a significant difference in the mechanism of initiation.
In bacteria, a 30S subunit binds directly to a ribosome-binding site. As a result, the initiation complex forms at a sequence surrounding the AUG initiation codon. When the mRNA is polycistronic, each coding region starts with a ribosome-binding site.
In eukaryotes, small subunits first recognize the 5′ end of the mRNA, and then move to the initiation site, where they are joined by large subunits.
Initiation in eukaryotic cytoplasm uses AUG as the initiator. The initiator tRNA is a distinct species, but its methionine does not become formylated. It is called tRNAiMet. So the difference between the initiating and elongating Met-tRNAs lies solely in the tRNA moiety, with Met-tRNAi used for initiation and Met-tRNAm used for elongation.
A unique reaction of eEF2(e=>eukaryotic,EF=>elongation factor) is its susceptibility to diphtheria toxin. The toxin uses NAD (nicotinamide adenine dinucleotide) as a cofactor to transfer an ADPR moiety (adenosine diphosphate ribosyl) on to the eEF2. The ADPR-eEF2 conjugate is inactive in protein synthesis. The ADP-ribosylation is responsible for the lethal effects of diphtheria toxin. The reaction is extremely effective: a single molecule of toxin can modify sufficient eEF2 molecules to kill a cell.
Only 61 triplets are assigned to amino acids. The other three triplets are termination codons (or stop codons) that end protein synthesis. They have casual names from the history of their discovery. The UAG triplet is called the amber codon; UAA is the ochre codon; and UGA is sometimes called the opal codon.
Two stages are involved in ending translation. The termination reaction itself involves release of the protein chain from the last tRNA. The post-termination reaction involves release of the tRNA and mRNA, and dissociation of the ribosome into its subunits.
Termination codons are recognized by class 1 release factors (RF). In E. coli two class 1 release factors are specific for different sequences . RF1 recognizes UAA and UAG; RF2 recognizes UGA and UAA.
In eukaryotes, there is only one single class 1 release factor, called eRF.
The dissociation of the remaining components (tRNA, mRNA, 30S and 50S subunits) requires the factor RRF, ribosome recycling factor. This acts together with EF-G in a reaction that uses hydrolysis of GTP. Like the other factors involved in release, RRF has a structure that mimics tRNA, except that it lacks an equivalent for the 3′ amino acid-binding region
In eukaryotic cytosolic ribosomes, another small RNA is present in the large subunit. This is the 5.8S RNA. Its sequence corresponds to the 5′ end of the prokaryotic 23S rRNA.
The 3′ end of the 16S rRNA is directly involved in the initiation reaction by pairing with the Shine-Dalgarno sequence in the ribosome-binding site of mRNA
23S rRNA has peptidyl transferase activity
The reduced specificity at the last position is known as third base degeneracy=>wobbling.
tRNAs are processed from longer precursors.
One feature that is common to all tRNAs is that the three nucleotides at the 3′ terminus, always the triplet sequence CCA, are not coded in the genome, but are added as part of tRNA processing.
The 5′ end of tRNA is generated by a cleavage action catalyzed by the enzyme ribonuclease P.
The addition of CCA is the result solely of an enzymatic process, that is, the enzymatic activity carries the specificity for the sequence of the trinucleotide, which is not determined by a template.
tRNAs are charged with amino acids by synthetases.
All synthetases function by the two-step mechanism.
First, the amino acid reacts with ATP to form aminoacyl~adenylate, releasing pyrophosphate. Energy for the reaction is provided by cleaving the high energy bond of the ATP.
Then the activated amino acid is transferred to the tRNA, releasing AMP.
The synthetases sort the tRNAs and amino acids into corresponding sets. Each synthetase recognizes a single amino acid and all the tRNAs that should be charged with it.
Synthetases use proofreading to improve accuracy.
A correctly paired aminoacyl-tRNA is able to make stabilizing contacts with rRNA. An incorrectly paired aminoacyl-tRNA does not make these contacts, and therefore is able to diffuse out of the A site.
Streptomycin inhibits protein synthesis by binding to 16S rRNA and inhibiting the ability of EF-G to catalyze translocation. It also increases the level of misreading of the pyrimidines U and C (usually one is mistaken for the other, occasionally for A). The site at which streptomycin acts is influenced by the S12 protein; the sequence of this protein is altered in resistant mutants.
The end of chapter four.
Chapter 5 Protein localization
Proteins are synthesized in two types of location:
The vast majority of proteins are synthesized by ribosomes in the cytosol.
A small minority are synthesized by ribosomes within organelles (mitochondria or chloroplasts).
Cytosolic (or "soluble") proteins are not localized in any particular organelle. They are synthesized in the cytosol, and remain there.
Most of the proteins in cytoplasmic organelles are synthesized in the cytosol and transported specifically to (and through) the organelle membrane,
The cytoplasm contains a series of membranous bodies, including endoplasmic reticulum (ER), Golgi apparatus, endosomes, and lysosomes. This is sometimes referred to as the "reticuloendothelial system." Proteins that reside within these compartments are inserted into ER membranes, and then are directed to their particular locations by the transport system of the Golgi apparatus.
Proteins that are secreted from the cell are transported to the plasma membrane and then must pass through it to the exterior.
Let’s check these animations:
http://www.sumanasinc.com/webcontent/anisamples/molecularbiology/proteinsecretion_fla.html
http://www.soest.hawaii.edu/%7Effd/bio275/movies/protein.sorting/protein.sorting.swf
http://www.soest.hawaii.edu/%7Effd/bio275/movies/protein.sorting/pd.1pass.swf
http://www.muskingum.edu/~brianb/CellPhys/Lect8/sld001.htm
http://www.abdn.ac.uk/sms/ugradteaching/BI2004/BI2004_14102005_9.ppt#2
http://www.abdn.ac.uk/sms/ugradteaching/BI2004/BI2004_11102005_7.ppt
http://academic.brooklyn.cuny.edu/biology/bio4fv/page/er_funct.htm
Protein degradation
http://www.kva.se/KVA_Root/pictures/prizes/nobel04/animation.html
http://www.sumanasinc.com/webcontent/anisamples/molecularbiology/lifecycleprotein_fla.html
http://www.rpi.edu/dept/bcbp/molbiochem/MBWeb/mb2/part1/protease.htm
Let’s check the details of transcription again:
Transcription
http://tidepool.st.usm.edu/crswr/protsynthmov.html
promoter structure
http://www.devbio.com/article.php?ch=5&id=38
DNA footprinting
http://users.rcn.com/jkimball.ma.ultranet/BiologyPages/F/Footprinting.html
Sigma Factor
http://binfo.ym.edu.tw/mb/pro_tc/sigma.htm
http://latin.arizona.edu/~mgen/micgen_98/Lect23/Lect_23.htm
Sporulation :
http://www.biology.ualberta.ca/facilities/multimedia/uploads/testing/spore2.swf
http://www.cat.cc.md.us/courses/bio141/lecguide/unit1/prostruct/sporegerm_an.html
http://info.bio.cmu.edu/Courses/03441/TermPapers/96TermPapers/spore/TOC.html
Terminator
http://www.phschool.com/science/biology_place/biocoach/transcription/procodgn.html
There are two types of terminators in E. coli.
Core enzyme can terminate in vitro at certain sites in the absence of any other factor. These sites are called intrinsic terminators.
Rho-dependent terminators are defined by the need for addition of rho factor (ρ) in vitro; and mutations show that the factor is involved in termination in vivo.
The end of chapter five.
Chapter 6 Operon
A group of genes used to control certain kind of cell function.
Lactose operon
http://highered.mcgraw-hill.com/olc/dl/120070/bio09.swf
http://highered.mcgraw-hill.com/olc/dl/120070/bio10.swf
http://highered.mcgraw-hill.com/olc/dl/120080/bio26.swf
http://highered.mcgraw-hill.com/olc/dl/120080/bio27.swf
http://highered.mcgraw-hill.com/olc/dl/120080/bio31.swf
http://bcs.whfreeman.com/thelifewire/content/chp13/1302001.html
Tryptophan operon
http://bcs.whfreeman.com/thelifewire/content/chp13/1302002.html
The end of chapter six.
Chapter 7 Gene Regulation
Regulatory RNA
http://www.massgeneral.org/pubaffairs/releases/110100ruvkun.htm
http://highered.mcgraw-hill.com/sites/0072437316/student_view0/chapter18/animations.html
http://web.mit.edu/bioedgroup/HBCF/animations.htm
http://faculty.plattsburgh.edu/donald.slish/Attenuation.html
http://www.microbelibrary.org/microbelibrary/files/ccImages/Articleimages/sandrin/trpoperon6.htm
http://jonathan.visick.faculty.noctrl.edu/260/week08.htm
http://users.rcn.com/jkimball.ma.ultranet/BiologyPages/P/Promoter.html
http://oregonstate.edu/instruction/bb492/lectures/Regulation.html
http://web.mit.edu/esgbio/www/pge/lac.html
http://wine1.sb.fsu.edu/bch5425/lect16/lect16.htm
http://web.indstate.edu/thcme/mwking/gene-regulation.html#euk
http://www.dls.ym.edu.tw/lesson/ige.htm
Stringent response
Deprivation of any one amino acid, or mutation to inactivate any aminoacyl-tRNA synthetase, is sufficient to initiate the stringent response.
The trigger that sets the entire series of events in train is the presence of uncharged tRNA in the A site of the ribosome.
when there is no aminoacyl-tRNA available to respond to a particular codon, the uncharged tRNA becomes able to gain entry. Of course, this blocks any further progress by the ribosome; and it triggers an idling reaction.
The components involved in producing (p)pGpp via the idling reaction have been identified through the existence of relaxed (rel) mutants. rel mutations abolish the stringent response,
The most common site of relaxed mutation lies in the gene relA, which codes for a protein called the stringent factor.
Ribosomes obtained from stringent bacteria can synthesize ppGpp and pppGpp in vitro, provided that the A site is occupied by an uncharged tRNA specifically responding to the codon. Ribosomes extracted from relaxed mutants cannot perform this reaction; but they are able to do so if the stringent factor is added.
The stringent factor (RelA) is an enzyme that catalyzes the synthetic reaction in which ATP is used to donate a pyrophosphate group to the 3′ position of either GTP or GDP. The formal name for this activity is (p)ppGpp synthetase.
How is ppGpp removed when conditions return to normal? A gene called spoT codes for an enzyme that provides the major catalyst for ppGpp degradation. The activity of this enzyme causes ppGpp to be rapidly degraded, with a half-life of ~20 sec.
The RelA enzyme uses GTP as substrate more frequently, so that pppGpp is the predominant product. However, pppGpp is converted to ppGpp by several enzymes; among those able to perform this dephosphorylation are the translation factors EF-Tu and EF-G. The production of ppGpp via pppGpp is the most common route, and ppGpp is the usual effector of the stringent response.
How does the state of the ribosome control the activity of RelA enzyme? Originally called relC, which turns out to be the same as rplK, which codes for the 50S subunit protein L11. This protein is located in the vicinity of the A and P sites.
Conformational change in this protein or some other component could activate the RelA enzyme, so that the idling reaction occurs instead of polypeptide transfer from the peptidyl-tRNA.
One round of (p)ppGpp synthesis is associated with release of the uncharged tRNA from the A site, so that synthesis of (p)ppGpp is a continuing response to the level of uncharged tRNA.
Entry of uncharged tRNA triggers the synthesis of a (p)ppGpp molecule, and the resulting expulsion of the uncharged tRNA allows the situation to be reassessed.
What does ppGpp do? Its primary regulatory effect is exerted by interacting directly with RNA polymerase. The most important consequence of this reaction is to inhibit initiation of transcription at certain promoters. There is also a general effect of reducing the rate of elongation of transcription, which results from increased pausing by RNA polymerase.
Let’s check these animations:
http://www.biochem.northwestern.edu/holmgren/Glossary/Definitions/Def-S/stringent_response.html
http://www.bmb.psu.edu/courses/micro401/Ch13Nts.htm
http://www.md.huji.ac.il/depts/humangenetics/glaser/glaser.html
Translation control
Protein binding(r-proteins—autogenous control) and RNA secondary structure(try operon--attenuation)
http://www.mun.ca/biochem/courses/3107/Topics/Trp_operon.html
http://www.ndsu.nodak.edu/instruct/mcclean/plsc431/prokaryo/prokaryo3.htm
http://web.mit.edu/esgbio/www/pge/pgeother.html
http://web.mit.edu/esgbio/www/pge/pgeother.html
RNA interference
http://fig.cox.miami.edu/~cmallery/150/gene/how_siRNA_works.htm
http://fig.cox.miami.edu/~cmallery/150/gene/siRNA.htm
http://www.sciscape.org/news_detail.php?news_id=751
http://www.nature.com/nrg/journal/v2/n2/animation/nrg0201_110a_swf_MEDIA1.html
http://www.promega.com/enotes/features/0305/RNAi.swf
http://www.pbs.org/wgbh/nova/sciencenow/3210/02.html
The end of chapter seven.
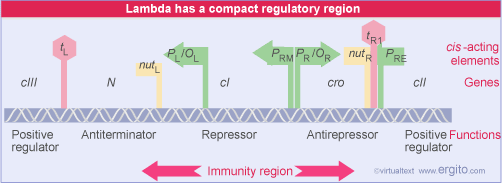
Chapter 8 Lambda Phage(http://life.nthu.edu.tw/~lslty/AMB/lec03-1-1.doc)
Bacteriophage Lambda: A Complex Viral Operon
Bacteriophage lambda is the prototype of a group of phages that have a very interesting "lifestyle." On the one hand, they can infect a cell and redirect the cell to become a factory for the production of new virus particles, resulting in the lysis of the cell. On the other hand, lambda can infect a cell, direct the integration of its genome into the DNA of the host, and reside there, replicating as a part of the chromosome, until such time as it activates and produces new virus particles and lyses the cell.
The first of these is called the lytic cycle, the second is the lysogenic cycle. Phage such as lambda that have these two possibilities are called temperate phage.
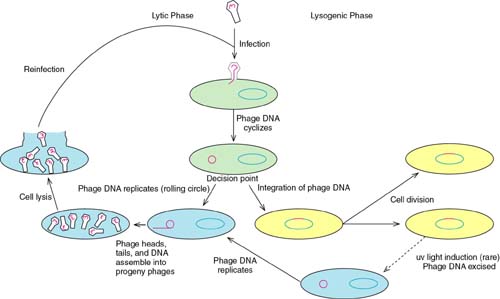
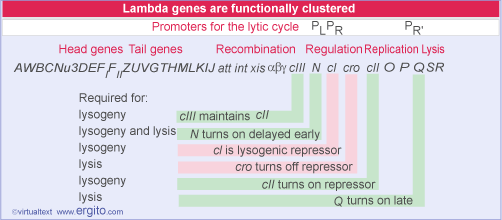
Phage gene expression during these two cycles uses the host RNA polymerase. But, rather than expressing new sigma factors or an entirely new polymerase , lambda uses operator-controlled promoters (operon type control) along with a type of regulation of transcription, antitermination.
The table summarizes the regulatory events during the lambda infectious cycle:
|
Event |
Lambda Gene Expressed |
Comment |
|
Initial infection |
cro, N |
Only N and cro are synthesized until the decision point is reached |
|
Lytic pathway |
cro, N, Q, late genes |
cro predominates at operators, N and Q are antiterminators |
|
Lysogenic pathway |
cI, cII, cIII, int |
cII and cIII collaborate to establish cI synthesis; after genome integration, only cI is expressed during maintenance of lysogeny. |
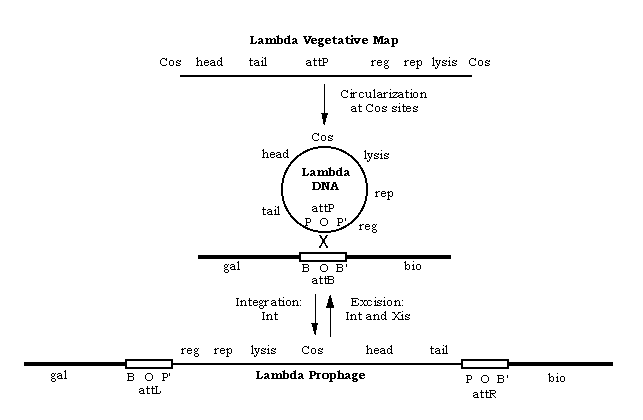
The lambda genome presented in a linear area (the way the DNA exists in the intact phage) and as a circle (the way the DNA forms after it enters the host cell):
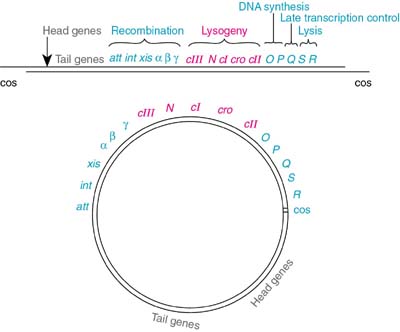
Lambda: The Initial Infection and the Lytic Pathway
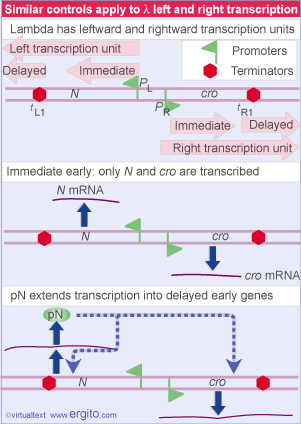
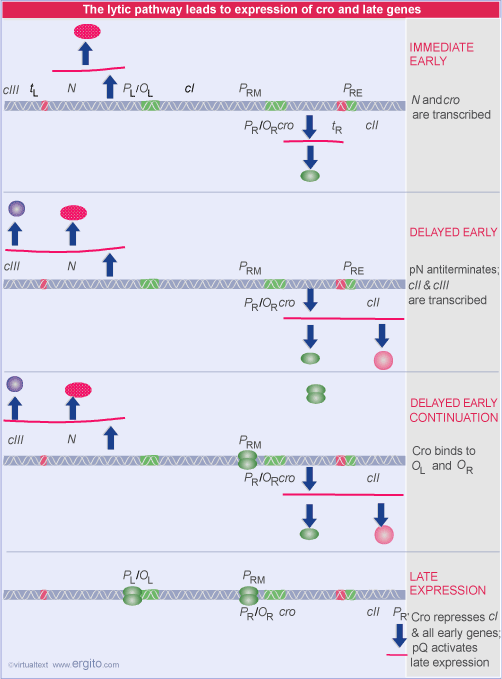
Immediately after lambda DNA enters the host cell, the host RNA polymerase begins to transcribe the lambda genome from two promoters: the PL (leftward) promoter and the PR (rightward) promoter. These two transcripts encode two proteins; cro (the abbreviation is a lambda jargon for "control of repressor and other things) and N. The phase is called immediate early transcription.

If the "decision" is made that this will be a lytic pathway then the delayed early genes are made next.

As the lytic infection proceeds, all of the phage late genes are expressed, including all of the proteins necessary to form the phage head and tail.

In order to switch from immediate early to delayed early, there is no change in the sigma factor or in the polymerase itself. Instead, lambda causes antitermination to occur at both rho-dependent and rho-independent termination sites. The antitermination is caused by two proteins, N and Q.
The effect of N on transcription from one of the two promoters makes the immediate early mRNAs.

In the presence of N, the transcription that was terminating at the site indicated in red, now continues through, producing a polycistronic mRNA. The same thing happens for transcription from the rightward promoter, PR. As a result, a number of gene products are made, not just N and cro. One of these is Q, another antiterminator that eventually opens up full transcription of all the late genes.
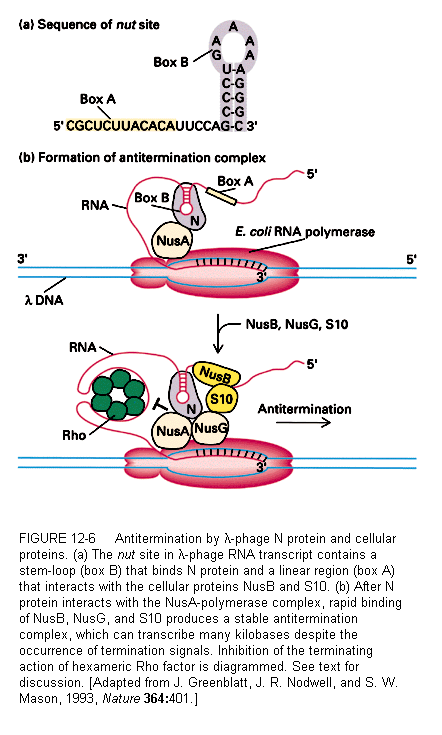
N acts in conjunction with four host cell proteins and the RNA polymerase. There is a site in the transcript RNA called the N-utilization site (nut site) to which N binds. Once bound to the region called Box B (a region of stem-loop structure), N can interact the polymerase through a protein called NusA (originally called the N-utilization substance A). Finally three other host proteins (NusB, NusG, and S10) bind at Box A of the nut site.
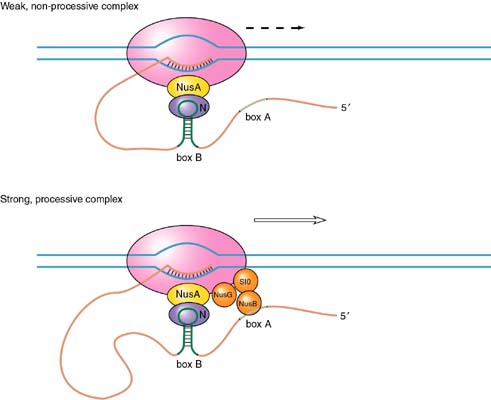
One model for the binding of N is that it prevents the pause of the polymerase that characterizes termination events. The stem-loop structure of the nut site Box B is some distance from the actual stem-loop at the termination site.
The other antiterminator, Q, acts in a very different manner.
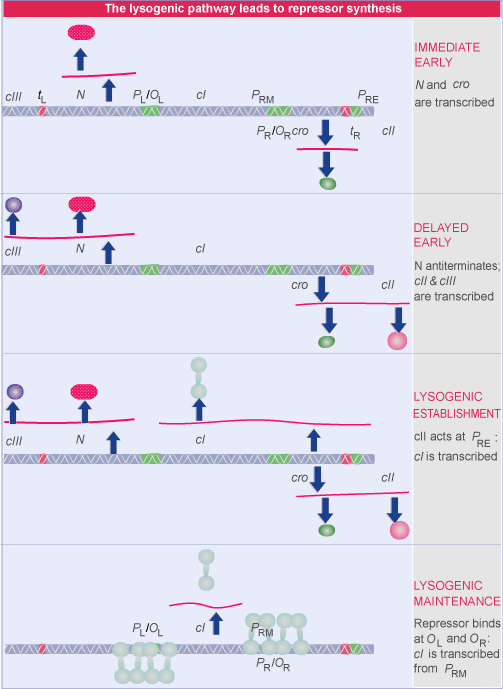
Lysogeny: Establishment and Maintenance
The lambda repressor is called cI ("c-one"). The origin of the names for the three genes originally identified as involved in lysogeny (cI, cII, and cIII) derives from the fact that mutants in these genes produced phage plaques on lawns of susceptible cells that were clear rather than the normal cloudy. (Plaques are clear areas where the virus has lysed the host cells).
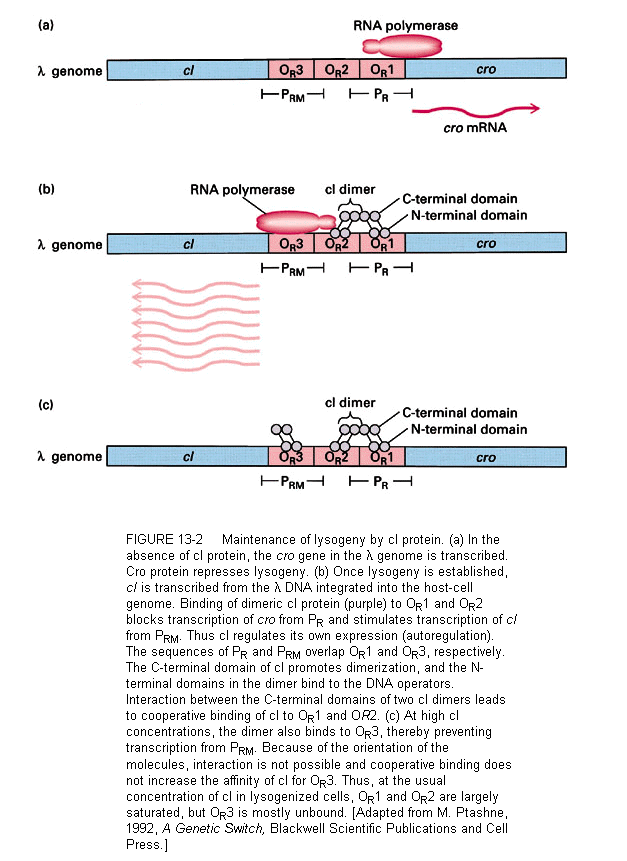
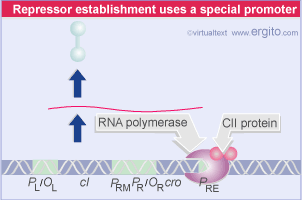
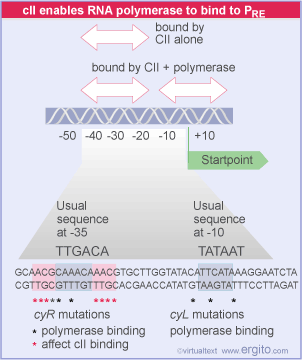
cI is a typical repressor protein, its function is to bind to operator sequences and prevent transcription. During the initial steps that establish lysogeny, cI must be made. The promoter for cI mRNA transcription is called PRE (promoter for repressor establishment).
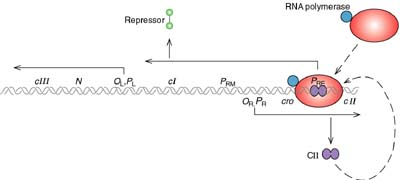
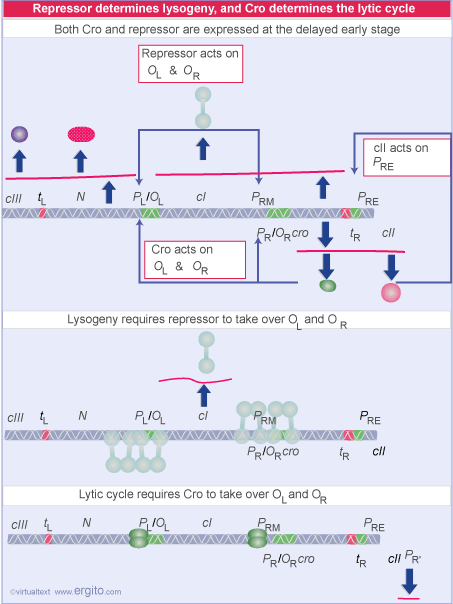
This promoter is unusual and the host RNA polymerase will not transcribe it without help. cII phage protein causes the polymerase to bind to PRE and transcribe mRNA for cI. cI mRNA is transcribed in the leftward direction from PRE. This means that the mRNA contains sequences that are complementary to cro mRNA (so-called "antisense" RNA). cII itself must be protected from proteolytic degradation by a host enzyme called HflA. This protection is afforded by the phage protein cIII.
When everything is right, cI is made and lysogeny is established. During this time the phage genome integrates into the host chromosome, catalyzed by a phage enzyme called integrase. In the lysogenic state, the phage genome is maintained in a quiescent state, expressing only one mRNA, the mRNA for cI, the repressor. Transcription of this mRNA now occurs from the promoter PRM (for "receptor maintenance").

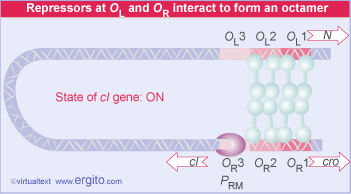
The lambda repressor binds to DNA as a dimer. There are two operator sites, OR and OL. Binding at OR and OL sites cuts of transcription of N and cro. A closer look at OR reveals that the operator region can be subdivided into three operators, OR1, OR2, and OR3. In fact, cI binds to both OR1 and OR2, and binding to OR2 actually stimulates the transcription of the cI mRNA.
In this sense, cI acts as both a positive and negative regulator of gene expression. cI negatively regulates cro and it positively regulates its own transcription.
cI binds to the three operator regions in a order of preference. It binds most tightly to OR1, next to OR2, and lastly to OR3. This order of binding will be critical to understanding the decision between lytic and lysogenic pathways.
Once lysogeny is established, the lambda DNA has integrated into the chromosome, the only viral protein being expressed is cI, and the viral DNA (now called a prophage) replicates along with its host. In fact, if a cell in this state (called a lambda lysogen) is subsequently infected by another lambda, nothing will happen. The second virus (called the super-infecting virus) is unable to make anything but the cI protein.
Lambda Wars: A Battle Between cI and Cro Determines Lysogenic or Lytic Pathways
During the earliest stage of infection with lambda, when the "decision point" is being reached (a reaction to the conditions), a competition takes place between cro and cI
cro is made right away, so are the two proteins cII (from PR) and cIII (from PL) after antitermination by N takes place. So, early on, both cro and cI may be present at some level.
What happens? The answer seems to be found at the OR operator. Both cI and cro bind to this OR operator. However, they bind differently. The binding of cro and cI to this region differs in terms of the order of interaction. The order is as follows:
cI: OR1, then OR2, then OR3
cro: OR3, then OR2, then OR1

If cI happens to get to its preferred binding sites first, it captures the operator, prevents cro synthesis and stimulates its own synthesis. However, if cro gets to its preferred site first, it captures the operator and prevents synthesis of cI (it does not stimulate its own synthesis, however).
What can lead to different levels of cro and cI? One answer is the environmental state of the cell. Two examples will help understand this.
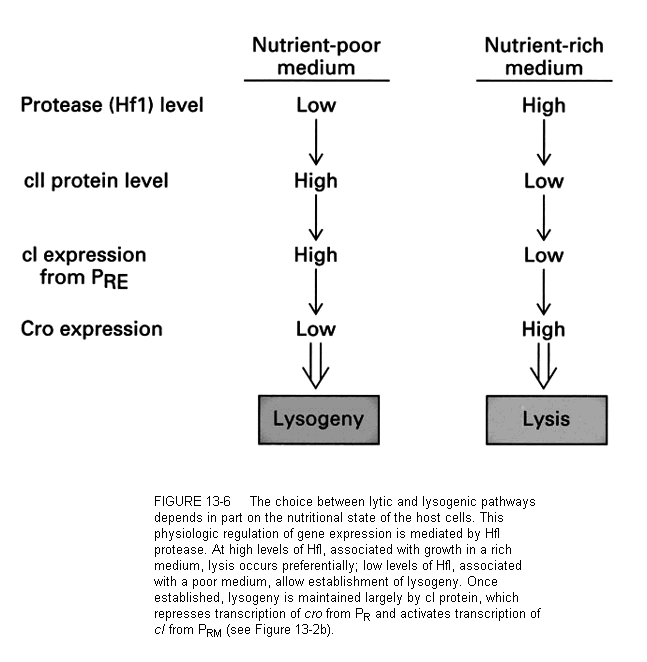
First, when the cell undergoes severe DNA damage from radiation (say, ultraviolet light), a repairs system is triggered called the SOS response. The SOS response activates a protease called the recA protease. This enzyme can destroy the lambda repressor cI. This means that cro gets a chance to win. Therefore under this condition, the phage goes into the lytic pathway, even if it has been in the lysogenic state. We say that UV induces the phage to the lytic pathway.

A second example involves the nutritional state of the cell. Remember from above that the cII protein is required for the host RNA polymerase to transcribe the cI mRNA from the promoter PRE. Lambda protein cIII protects cII from destruction by the cellular protease, HflA.
This protease is sensitive to the glucose level in the environment of the cell. Low glucose results in high cAMP. The HflA protease is activated by high cAMP and is able to overcome the inhibition by cIII. Therefore, when the cell is in a poor nutritional state (signaled by low glucose) the lytic pathway is favored over the lysogenic.
These two examples show that lambda has evolved to make the lytic vs. lysogenic "decision" based upon the fitness of the cell as a long term host. If the cell is healthy (e.g., no DNA damage or in good nutritional state) the virus can make a home for its genome there (lysogeny). When the state of the cell does not look good for the long term (UV damage or poor growth medium) the virus replicates quickly, making progeny and lysing the cell.
Let’s review the whole concept:

(http://life.nthu.edu.tw/~lslty/AMB/lec03-1-1.doc )
Or

(http://www.science.siu.edu/microbiology/micr460/460%20Pages/Lambda.map.html )
Let’s check this animation:
http://www.blackwellpublishing.com/trun/artwork/Animations/Lambda/lambda.swf

(http://www.accessexcellence.org/RC/VL/GG/bact_Lambda.html
http://www-micro.msb.le.ac.uk/3035/Phages.html#Lambda



Binding specificity for CI: OR3 < OR2 < OR1 and OL1 > OL2 > OL3
Binding specificity for Cro: OR3 > OR2 ~ OR1 and OL1 ~ OL2 < OL3



http://www.accessexcellence.org/RC/VL/GG/bact_Lambda.html
http://www.science.siu.edu/microbiology/micr460/460%20Pages/460.lambda.html
http://www.acsu.buffalo.edu/~koudelka/Bacteriophage_lambda_lecture%201.ppt#1
http://www.csun.edu/~hcbio027/biotechnology/lec4a/lambda.html
http://www.blackwellpublishing.com/trun/artwork/Animations/Lambda/lambda.swf
http://www.biology.ualberta.ca/facilities/multimedia/uploads/microbiology/lambda.swf
http://www-biology.ucsd.edu/classes/bimm100.WI00/XIV.Lambda.html
http://escience.ws/b572/L16a/L16a.htm
http://biology.fullerton.edu/biol302/lambdaweb/
http://home.earthlink.net/~dayvdanls/lambdavideo.htm
http://faculty.uml.edu/mark_hines/81.201/media/chapter_17.ppt
http://bioweb.wku.edu/courses/biol566/L2lambda.html
When lambda DNA enters a new host cell, the lytic and lysogenic pathways start off the same way. Both require expression of the immediate early and delayed early genes. But then they diverge: lytic development follows if the late genes are expressed; lysogeny ensues if synthesis of the repressor is established.
Lambda has only two immediate early genes, transcribed independently by host RNA polymerase:
N codes for an antitermination factor whose action at the nut sites allows transcription to proceed into the delayed early genes (see Antitermination requires sites that are independent of the terminators).
cro has dual functions: it prevents synthesis of the repressor (a necessary action if the lytic cycle is to proceed); and it turns off expression of the immediate early genes (which are not needed later in the lytic cycle).
The cII-cIII pair of regulators is needed to establish the synthesis of repressor.
The Q regulator is an antitermination factor that allows host RNA polymerase to transcribe the late genes.
(http://www.mgm.ufl.edu/~gulig/id/04phages.pdf)
Two immediate early genes, N and cro, are transcribed by host RNA polymerase. N is transcribed toward the left, and cro toward the right. Each transcript is terminated at the end of the gene. pN is the regulator that allows transcription to continue into the delayed early genes. It is an antitermination factor that suppresses use of the terminators tL and tR
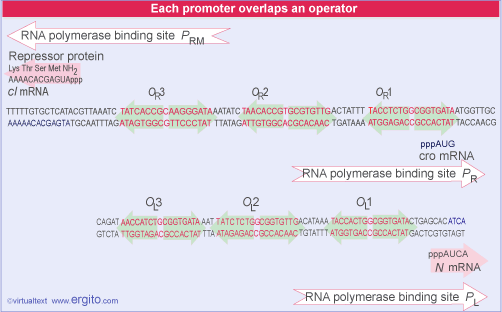
That OL1 and OR1 lie more or less in the center of the RNA polymerase binding sites of PL and PR, respectively. Occupancy of OL1-OL2 and OR1-OR2 thus physically blocks access of RNA polymerase to the corresponding promoters.
A different relationship is shown between OR and the promoter PRM for transcription of cI. The RNA polymerase binding site is adjacent to OR2. This explains how repressor autogenously regulates its own synthesis. When two dimers are bound at OR1-OR2, the dimer at OR2 interacts with RNA polymerase
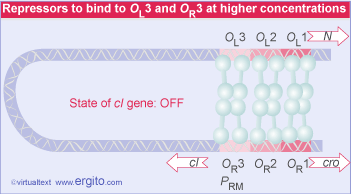
Mutations in the host genes hflA and hflB increase lysogeny—hfl stands for high frequency lysogenization. The mutations stabilize cII because they inactivate host protease(s) that degrade it.
The end of chapter eight.
Chapter 9 DNA Replication
Let’s check these animations:
http://highered.mcgraw-hill.com/sites/0072437316/student_view0/chapter14/animations.html#
http://www.fed.cuhk.edu.hk/~johnson/teaching/genetics/animations/dna_replication.htm
http://www.johnkyrk.com/DNAreplication.html
http://big.mcw.edu/display.php/724.html
http://www.bst.ntu.edu.tw/curriculum/Mobi%20WAY/Replication4.ppt#1
Try this
http://www.pbs.org/wgbh/nova/genome/sequencer.html#
Conjugation:
http://tidepool.st.usm.edu/crswr/bactconjug.html
http://www.emunix.emich.edu/~rwinning/genetics/bactrec2.htm
http://oregonstate.edu/instruction/bb492/lectures/DNAI.html
http://info.bio.cmu.edu/Courses/03441/TermPapers/97TermPapers/mutator/struct.htm
http://chem-faculty.ucsd.edu/kraut/bpol.html
http://www.biosci.ohio-state.edu/~mgonzalez/Micro521/04.html
http://138.192.68.68/bio/Courses/biochem2/DNA/DNAPolymerase.html
http://veghome.ucdavis.edu/bis101/ANIMATIO/DNAREPCT.DIR
http://www.ncc.gmu.edu/dna/replicat.htm
What feature of a bacterial (or plasmid) origin ensures that it is used to initiate replication only once per cycle? oriC contains 11 copies of the sequence , which is a target for methylation at the N6 position of adenine by the Dam methylase. Before replication, the palindromic target site is methylated on the adenines of each strand. Replication inserts the normal (nonmodified) bases into the daughter strands, generating hemimethylated DNA, in which one strand is methylated and one strand is unmethylated. So a hemimethylated origin cannot be used to initiate a replication cycle.
What is responsible for the delay in remethylation at oriC and dnaA? The most likely explanation is that these regions are sequestered in a form in which they are inaccessible to the Dam methylase. SeqA binds to hemimethylated DNA more strongly than to fully methylated DNA. and then its continued presence prevents formation of an open complex at the origin.
The end of chapter nine.
Chapter 10 Recombination and Repair
There are three types of recombination:
Homologous recombination, Site-specific recombination, Transposition(transposome and RNA virus RNA polymerase jumping---copy choice included).
Let’s check tnis animation:
http://www.web-books.com/MoBio/Free/Ch8D.htm
Recombination needs enzymes to function:
DNA topoisomerase
http://www.jonathanpmiller.com/intercalation/topoisomerase.htm
http://www.maich.gr/natural/staff/sotirios/topo.html
http://cmgm.stanford.edu/biochem201/Handouts/DNAtopo.html
http://biology.kenyon.edu/BMB/Chime/topo1/frames/topoitxt.htm
DAN repair system:
http://phys.thu.edu.tw/~ctshih/teach/biophys/DNA%20damage%20and%20repair.ppt#1
http://www.nature.com/nrc/journal/v1/n1/animation/nrc1001-022a_swf_MEDIA1.html
http://www.science.siu.edu/microbiology/micr460/460%20Pages/460.DNArepair.html
http://www.csu.edu.au/faculty/health/biomed/subjects/molbol/basic1.htm
http://www.mun.ca/biochem/courses/3107/Topics/Repair.html
http://www.sparknotes.com/biology/molecular/dnareplicationandrepair/section3.rhtml
http://www.funpecrp.com.br/gmr/year2003/vol1-2/sim0001_full_text.htm
http://www-biology.ucsd.edu/classes/bimm100.FA00/15.DNArepair.html
http://en.wikipedia.org/wiki/SOS_response
Transposons Retroviruses and retroposons:
http://en.wikipedia.org/wiki/Transposon
http://nucleus.cshl.org/protarab/TnAnnotation.htm
hybrid dysgenesis
http://www.mun.ca/biology/scarr/P-element_hybrid_dysgenesis.htm
http://opbs.okstate.edu/~melcher/MG/MGW3/MG32217.html
http://engels.genetics.wisc.edu/Pelements/Pt.html
(http://www.ndsu.nodak.edu/instruct/mcclean/plsc431/transelem/trans1.htm Ac/Ds)
http://www.whfreeman.com/kuby/content/anm/kb03an01.htm
http://www.galaxygoo.org/hiv/hiv_lifecycle.html
http://www-micro.msb.le.ac.uk/Video/HIV.mov
Retrovirus replication:
http://www.accessexcellence.org/RC/VL/GG/retrovirus.html
http://www.mun.ca/biochem/courses/3107/Topics/retrovirus_replication.html
http://www.mun.ca/biochem/courses/3107/images/Stryer/Stryer_F32-29.jpg
http://www.mun.ca/biochem/courses/3107/images/MVH/MVH_fi24p45.gif
http://www-micro.msb.le.ac.uk/3035/Retroviruses.html
Oncogene:
http://atp.life.nctu.edu.tw/~biocenter/sections.php?op=viewarticle&artid=284
http://cmbi.bjmu.edu.cn/news/report/2004/Oncogene.htm
http://en.wikipedia.org/wiki/Oncogene
http://web.indstate.edu/thcme/mwking/oncogene.html
http://www.britannica.com/nobel/micro/438_45.html
http://outreach.mcb.harvard.edu/animations_S03.htm
http://www.learner.org/channel/courses/biology/units/cancer/images.html
http://science.education.nih.gov/supplements/nih1/cancer/activities/activity2_animations.htm
http://www.pbs.org/wgbh/nova/cancer/program.html
http://www.pbs.org/wgbh/nova/cancer/grow_flash.html
Ty element
http://www.ndsu.nodak.edu/instruct/mcclean/plsc431/transelem/trans2.htm
The end of chapter ten.